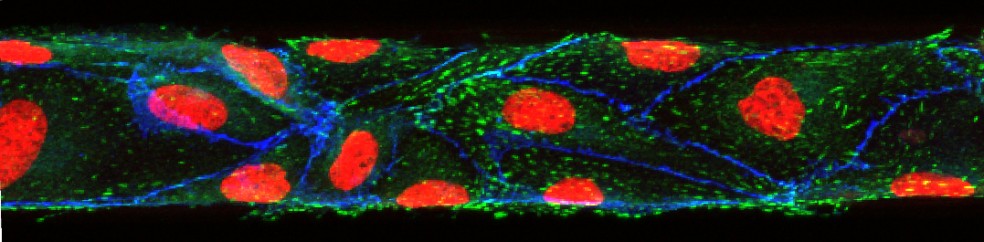
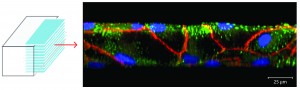
Microphysiologic Cell Culture Systems are microfluidic platforms on which multiple in vitro tissues can communicate with each other via soluble metabolites that re-circulate through a medium stream. The systems are also referred to as body-on-a-chip systems or micro cell culture analogs (µCCAs).
My research combines tissue engineering and microfabrication to construct microphysiological systems and to examine how cooperative interactions of organs result in the overall functioning of the human body. The collective effects of inter-organ metabolic exchanges are not only directly relevant to evaluating new drugs, they also raise a number of fascinating Systems Biology questions concerning the mechanisms of communication between organs that lead to the overall response of the human body to chemical and biological challenges.
Here is a paper that reviews some of the literature regarding micro physiological systems and outlines their possibilities for research:
Mandy B. Esch, Alec Smith, Jean-Mathew Prot, Charlotta Oleaga Sancho, James Hickman, Michael L. Shuler, How Multi-Organ Microdevices Can Help Foster Drug Development. Advanced Drug Delivery Reviews, 2014, 69/70, 158-169.
Here are two papers that present a micro physiological system with GI tract and liver tissues:
Mandy B. Esch, Gretchen J. Mahler, Michael L. Shuler, Body-on-a-Chip simulation with gastrointestinal and liver tissue suggests that ingested nanoparticles have the potential to cause liver injury. Lab on a Chip, 2014, 14, 3081-3092. doi: 10.1039/c41c00371c
Mahler, G.J., Esch M.B., Shuler, M.L. “Characterization of a Gastrointestinal Tract Microscale Cell Culture Analog Used to Predict Drug Toxicity ”, Biotechnol & Bioeng, 2009, 104(1), 193-205.
 Mahler, G.J., Esch M.B., Shuler, M.L. “Characterization of a Gastrointestinal Tract Microscale Cell Culture Analog Used to Predict Drug Toxicity ”, Biotechnology & Bioengineering, 2009, 104(1), 193-205.
Mahler, G.J., Esch M.B., Shuler, M.L. “Characterization of a Gastrointestinal Tract Microscale Cell Culture Analog Used to Predict Drug Toxicity ”, Biotechnology & Bioengineering, 2009, 104(1), 193-205.