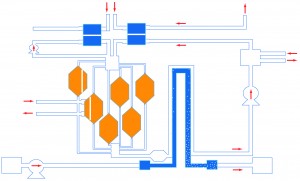
Microphysiologic Cell Culture Systems are microfluidic platforms on which multiple in vitro tissues can communicate with each other via soluble metabolites that re-circulate through a medium stream. The systems are also referred to as body-on-a-chip systems or micro cell culture analogs (µCCAs).
My research combines tissue engineering and microfabrication to construct microphysiological systems and to examine how cooperative interactions of organs result in the overall functioning of the human body. The collective effects of inter-organ metabolic exchanges are not only directly relevant to evaluating new drugs, they also raise a number of fascinating Systems Biology questions concerning the mechanisms of communication between organs that lead to the overall response of the human body to chemical and biological challenges.
Here is a paper that reviews some of the literature regarding micro physiological systems and outlines their possibilities for research:
Mandy B. Esch, Alec Smith, Jean-Mathew Prot, Charlotta Oleaga Sancho, James Hickman, Michael L. Shuler, How Multi-Organ Microdevices Can Help Foster Drug Development. Advanced Drug Delivery Reviews, 2014, in print, doi: 10.1016/j.addr.2013.12.003
————————————————————————————————————————————-
Body-on-a-chip systems
High quality, in vitro screening tools are essential in identifying promising compounds during drug development. Tests with currently used cell-based assays only provide an indication of a compound’s potential therapeutic benefits to the target tissue, but not the whole body. Data obtained with animal models often can not be extrapolated to humans. Multi-compartment microfluidic based devices, particularly those that are physical representations of physiologically based pharmacokinetic (PBPK) models, may contribute to improving the drug development process. These scaled down devices, called micro cell culture analogs (µCCAs) or “body-on-a-chip”, can simulate multi-tissue interactions. Here are two recent studies from our lab that utilize the body-on-a-chip concept:
Mandy B. Esch, Gretchen J. Mahler, Michael L. Shuler, Body-on-a-Chip simulation with gastrointestinal and liver tissue suggests that ingested nanoparticles have the potential to cause liver injury. (2014) Lab on a Chip, in print. doi: 10.1039/c41c00371c
Mahler, G.J., Esch M.B., Shuler, M.L. “Characterization of a Gastrointestinal Tract Microscale Cell Culture Analog Used to Predict Drug Toxicity ”, Biotechnol & Bioeng, 2009, 104(1), 193-205
————————————————————————————————————————————-
Microfabricated porous 3D membrane is tissue scaffold for on-chip GI tract epithelium
 A physiologically correct, on-chip, first pass metabolism model of oral absorption of drugs and environmental toxicants requires a microfluidic cell culture systems with three dimensional, multi cell-type cultures that replicate in vivo cell morphologies and structure. “First pass metabolism” refers to the absorption of ingested substances through intestinal tissue and their immediate transfer to the liver where enzymes can metabolize them before they enter systemic circulation. The process considerably decreases the bioavailability of ingested substances and is of interest to drug developers, toxicologists, and nutrition scientists. With such a system we can determine subtle physiological effects of drugs and toxicants that go beyond gross toxicity (e.g. viability). We aim to incorporate 3-dimensional in vitro tissues in the model and validate its ultimate ability to simulate first pass metabolism in humans through comparing results obtained with in vitro devices using cell types from rats, and humans to those obtained with in vivo rat experiments.
A physiologically correct, on-chip, first pass metabolism model of oral absorption of drugs and environmental toxicants requires a microfluidic cell culture systems with three dimensional, multi cell-type cultures that replicate in vivo cell morphologies and structure. “First pass metabolism” refers to the absorption of ingested substances through intestinal tissue and their immediate transfer to the liver where enzymes can metabolize them before they enter systemic circulation. The process considerably decreases the bioavailability of ingested substances and is of interest to drug developers, toxicologists, and nutrition scientists. With such a system we can determine subtle physiological effects of drugs and toxicants that go beyond gross toxicity (e.g. viability). We aim to incorporate 3-dimensional in vitro tissues in the model and validate its ultimate ability to simulate first pass metabolism in humans through comparing results obtained with in vitro devices using cell types from rats, and humans to those obtained with in vivo rat experiments.
Here is a paper that describes the fabrication of a 3D tissue scaffold:
Esch, M.B., Sung, J.H., Yang, J. Yu, J., March, J.C., Shuler, M.L. “On Chip Porous Polymer Membranes for Integration of Gastrointestinal Tract Epithelium with Microfluidic ‘Body-on-a-Chip’ Devices”, Biomedical Microdevices, 2012, 14 (5), 895-906
————————————————————————————————————————————-
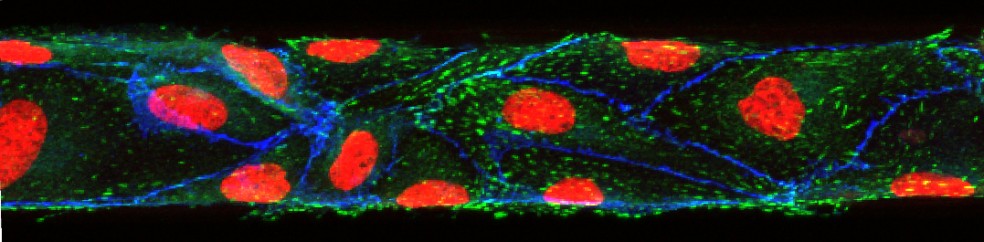
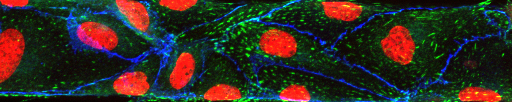
50 micrometer in vitro microvasculature (endothelial lining)
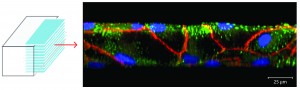 In vitro microvascular endothelial linings can be used to investigate biophysical and molecular mechanisms that play a role in circulatory disease phenomena such as inflammation, responses of the immune system and cancer cell attachment to the endothelium. In vivo, endothelial cells grow on the inner surface of blood vessels and are confined by its geometry. In the smallest vessels of the microvasculature, this confinement leads to a significant bend within each cell. To imitate these geometric constraints within an in vitro model of the endothelial lining, we have fabricated nearly round channels (70 µm wide, 50 µm high) and cultured human umbilical vein endothelial cells (HUVECs) within them. We have characterized the developed model and compared the results with those obtained with conventional microfluidic channels with square cross-sectional profiles. Our results indicate that shear stress is responsible for the conforming of HUVECs to the channel walls of square profile channels under fluid flow and cause the resulting square geometry of the in vitro endothelial lining.
In vitro microvascular endothelial linings can be used to investigate biophysical and molecular mechanisms that play a role in circulatory disease phenomena such as inflammation, responses of the immune system and cancer cell attachment to the endothelium. In vivo, endothelial cells grow on the inner surface of blood vessels and are confined by its geometry. In the smallest vessels of the microvasculature, this confinement leads to a significant bend within each cell. To imitate these geometric constraints within an in vitro model of the endothelial lining, we have fabricated nearly round channels (70 µm wide, 50 µm high) and cultured human umbilical vein endothelial cells (HUVECs) within them. We have characterized the developed model and compared the results with those obtained with conventional microfluidic channels with square cross-sectional profiles. Our results indicate that shear stress is responsible for the conforming of HUVECs to the channel walls of square profile channels under fluid flow and cause the resulting square geometry of the in vitro endothelial lining.
Here is a paper on the development of the in vitro endothelium:
Esch, M.B., Post, D., Shuler, M.L., Stokol, T. “Characterization of Small Diameter In Vitro Endothelial Linings of the Microvasculature,” Tissue Engineering A, 2011, 17, 2965-2971
————————————————————————————————————————————-
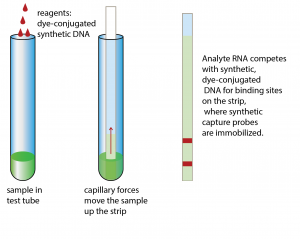
Paper-based microfluidics for 30 min RNA detection
Paper-based microfluidics are the basis for pregnancy test strips that detect hormones via antibodies. We employed the same principle to detect RNA of a pathogen (Cryptosporidium parvum). We immobilized synthetic DNA that is complementary to the RNA on the test strip. While the sample moves up the strip via capillary forces, the RNA in the sample comes into very close contact with the immobilized probe. In our assay, the RNA competes with a second dye-conjugated synthetic DNA with the same sequence that we initially added to the sample. If the RNA of the pathogen is present this synthetic DNA will not have a chance to bind. In this competitive assay format, no color on the strip means the pathogen RNA was present. To read more about this assay, check out our publication in Analytical Chemistry from 2001:
Esch, M.B.; Bäumner, A.; Durst, R.A. “Rapid Visual Detection of Viable Cryptosporidium parvum on Test Strips using Oligonucleotide-tagged Liposomes” Analytical Chemistry, 2001, 73(13), 3162-3167